





Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a Weed
October 27th, 2017
A new research progress recently published on Nature Communications titled “Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed” led by Professor Guo Longbiao from China National Rice Research Institute (CNRRI) of Chinese Academy of Agricultural Sciences (CAAS) with the collaboration of Professor Fan Longjiang from Zhejiang University. The research teams acquire a new understanding of the molecular mechanisms underlying the extreme adaptation and invasiveness of the hexaploid species Echinochloa crus-galli (E. crus-galli), a pernicious weed in agricultural fields worldwide.
E. crus-galli is one of the most serious agricultural weeds in the world. Barnyardgrass (E. crus-galli) and rice belong to Gramineae, both have similar biological characteristics like rapid growth rates, plant type, prolonged dormance and others. That is why, it is very difficult to control the paddy fields associated with Barnyardgrass via manual weeding. Paddy weeding is currently largely dependent on chemical herbicides. The use of a large number of herbicides not only pollutes the environment, but also increases the cost of production and resistance to Barnyardgrass. Therefore, the selection of "green rice" with inhibition of barnyardgrass is an important way to reduce the use of chemical herbicides.
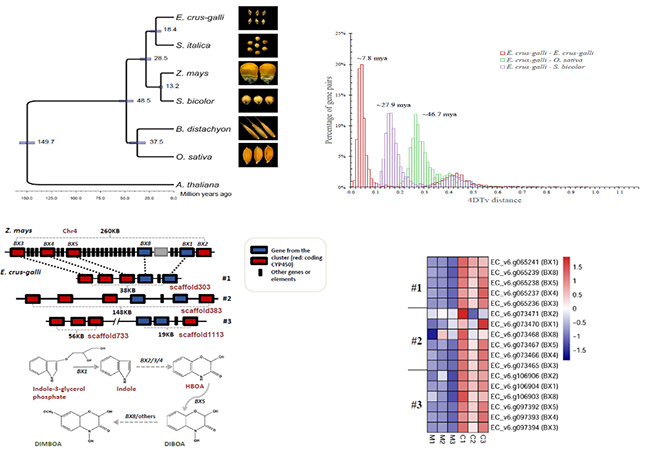
The genome of barnyardgrass was sequenced, and the defensive secondary metabolites of barnyard grass were synthesized by clustered genes, which were used to compete with rice and resist rice pathogen, and provide an important genetic resource for rice C4 breeding. An extremely large repertoire of genes encoding cytochrome P450 monooxygenases and glutathione S-transferases associated with detoxification are found. Two gene clusters involved in the biosynthesis of an allelochemical 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA) and a phytoalexin momilactone A are found in the E. crus-galli genome, respectively. The allelochemical DIMBOA gene cluster is activated in response to co-cultivation with rice, while the phytoalexin momilactone A gene cluster specifically to infection by pathogenic Pyricularia oryzae. The results provide a new understanding of the molecular mechanisms underlying the extreme adaptation of the weed.
Prof. Guo Longbiao of China National Rice Research Institute of CAAS, and Dr. Qiu Jie of Zhejiang University are the co-first authors of this study which is supported by the National Natural Science Foundation of China. Professor Fan Longjiang from Zhejiang University and Prof. Bai Lianyang of Hunan Academy of Agricultural Sciences are co-corresponding authors. This work was financed by State Key Lab of Rice Biology of China, Zhejiang Natural Science Foundation (LZ17C130001), National Natural Science Foundation of China (31401453), and so on. More details can be found in the link below: http://www.nature.com/articles/s41467-017-01067-5.
· FED:a web tool for foreign element detection of genome-edited organism
· The C2H2 zinc-finger protein LACKING RUDIMENTARY GLUME 1 regulates spikelet development in rice
· A strigolactones biosynthesis gene contributed to the Green Revolution in rice
· Natural variation in the promoter of TGW2 determines grain width and weight in rice
CNRRI Today
Copyright © 2014- China National Rice Research Institute