


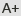



QTL qLSCHL4 increases yield of indica super rice 93-11
June 25th, 2014
The basic premise of high yield in rice is to improve leaf photosynthetic efficiency and coordinate the source-sink relationship in rice plants. In selection breeding, molecular markers become extremely popular among breeders and scientists. In China, significant progress has been achieved in gene exploration and genetic studies of yield-related traits in rice; a number of quantitative trait loci (QTLs) of important traits in rice are detected in different genetic populations. To date, nearly 20 yield-related QTLs have been isolated by scholars using map-based cloning methods , e.g., Gn1a, Ghd7, DEP1, NAL1 (SPIKE and GPS) and IPA1; these genes control plant architecture and panicle type, ultimately affecting the yield of rice mainly through pleiotropic effects.
Recently, researchers from China National Rice Research Institute (CNRRI) found a major quantitative trait locus (QTL) related to morphological traits and chlorophyll content of rice leaves at the stages of heading to maturity. This QTL (qLSCHL4) related to flag leaf shape and chlorophyll content was detected at both stages in recombinant inbred lines constructed using the indica rice cultivar 93-11 and the japonica rice cultivar Nipponbare. Map-based cloning and expression analysis showed that LSCHL4 is allelic to NAL1, a gene previously reported in narrow leaf mutant of rice. Overexpression lines transformed with vector carrying LSCHL4 from Nipponbare and a near isogenic line of 93-11 (NIL-9311) had significantly increased leaf chlorophyll content, enlarged flag leaf size, and improved panicle type.
The average yield of NIL-9311 was 18.70% higher than that of 93-11. These results indicate that LSCHL4 had a pleiotropic function. Exploring and pyramiding more high-yield alleles resembling LSCHL4 for super rice breeding provides an effective way to achieve new breakthrough in raising rice yield and generate new ideas for solving the problem of global food safety.
This work was supported by grants from the National Natural Science Foundation of China (31221004 and 91335105), and Provincial Science Fund for Distinguished Young Scholars of Zhejiang (R3100100). The research result was published in Mol. Plant (2014) doi:10.1093/mp/ssu055.
http://mplant.oxfordjournals.org/content/early/2014/06/11/mp.ssu055.full.pdf+html
Genetic linkage map of putative QTLs for flag leaf width and cholorphyll content at different growth stages in rice.
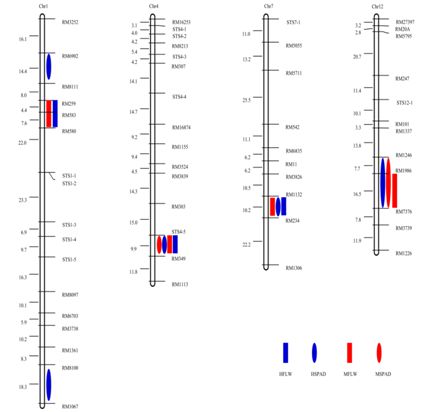
Comparison Morphology and Cross-Sections of Flag Leaves. (A) Characterization of large vascular bundle (LVB) and small vascular bundle (SVB) number of 93-11, NIL-9311, and NPB. Each column represents mean ± SD. Columns with different letters were significantly different (P < 0.01, least significant difference test). (B) Flag leave morphology of Nipponbare, 93-11, and NIL-9311 (bar=2.5cm). (C–E) Cross-sections of flag leaves stained with Safranin O and Fastgreen FCF in Nipponbare, 93-11, and NIL-9311. Bar=100μm.

Morphologies of Nipponbare (NPB) Plant and Overexpressor (ubi:qLSCHL4)
Transgenic Nipponbare Plant (NPB-OE). (A) Plant morphology (bar=30cm). (B) The culm cross-section of the fourth internode (bar=2mm). (C) Leaf shape (bar=5cm). (D) Panicle (bar=4cm) of Nipponbare (NPB) and overexpression Niponbare (NPB-OE).
· Study Reveals How the Clock Component OsLUX Regulates Rice Heading
· Scientists Further Unravelled the Underlying Mechanism of Heading Date Control in Rice
· Improving the efficiency of prime editing in rice
· A New Infection Mechanism of Ustilaginoidea virens is Revealed
· Scientists Further Reveal the Infection Mechanism of Ustilaginoidea virens
CNRRI Today
Copyright © 2014- China National Rice Research Institute