


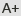



Robust genome editing of CRISPR-Cas9 at NAG PAMs in rice
January 29th, 2018
Recently, researchers from China National Rice Research Institute, CAAS and Institute of Genetics and Developmental Biology, CAS cooperatively reported that CRISPR-Cas9 system could effectively recognize NAG PAMs in rice on the paper entitled Robust genome editing of CRISPR-Cas9 at NAG PAMs in rice. The work is publicly available online on SCIENCE CHINA Life Sciences (SCLS). Simultaneously, Holger Puchta’s commentary on the work entitled Broadening the applicability of CRISPR/Cas9 in plants was published by SCLS, in which the theoretical value and application prospect of the above research was highlighted.
Genome editing has become one of the revolutionary breakthroughs in life science. As the most well-developed and pervasive genome editing system, CRISPR-Cas9 has been applied extensively in the animal, plant and microorganism. However, the range of selectable targets is constrained by a combination of nucleotides, NGG. Many previous studies tried to exceed the limitation by employing Cas9 variants and analogous protein and achieved a series of accomplishments, like VQR/VRER or Cpf1 that was successfully applied in rice by Wang’s group.
Previous studies in bacterial and human cells revealed that SpCas9 can recognize NAG PAM, but its efficiency is only about a fifth of NGG PAM sites, which hinders its application in genome editing. Researchers selected five targets, which have two sites in rice genome and include these two kinds of PAM (NGG and NAG) by bioinformatics methods. The results revealed that CRISPR-Cas9 system could effectively recognize NAG PAM, and its efficiency is about 76.44% of that of NGG. Besides, a comparison between two PAMs reveals that the off-target effect of NAG shows relatively low tolerance of mismatching. Among 8 targets testing in rice protoplast, the efficiency ranges from 0.95% to 8.45%. However, the efficiency ranges from 15.52% to 82.14% (43.15% on average) during stable transformation. Further investigation shows that NGG and NAG could be recognized simultaneously.
This work expands the range of genome editing with CRISPR-Cas9 system and alerts researchers to the potential off-target with NAG PAM in its conventional use.
This work is supported by the National Key Research and Development Program of China (2016YFD0101800), the National Natural Science Foundation of China (91635301), the Zhejiang Provincial Natural Science Foundation of China (LZ14C130003), and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences. Xixun Hu from Wang’s group and Xiangbing Meng from Li’s group are co-first authors. More details are available on the link below: https://link.springer.com/article/10.1007/s11427-017-9247-9
· Study Reveals How the Clock Component OsLUX Regulates Rice Heading
· Scientists Further Unravelled the Underlying Mechanism of Heading Date Control in Rice
· Improving the efficiency of prime editing in rice
· A New Infection Mechanism of Ustilaginoidea virens is Revealed
· Scientists Further Reveal the Infection Mechanism of Ustilaginoidea virens
CNRRI Today
Copyright © 2014- China National Rice Research Institute