


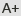



Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems
December 11st, 2018
Recently, the innovation team of rice chromosome engineering and genome editing from China National Rice Research Institute (CNRRI) of Chinese Academy of Agricultural Sciences (CAAS) published a research paper entitled Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems on Science China Life Sciences. In the paper, researchers developed a simplified and lowcost strategy that combines common experiment kits for NGS library construction and a user-friendly web tool for high-throughput analysis of multiple samples at multiple target sites. The work will facilitate the application of NGS sequencing in mutation identification, particularly for laboratories that do not have researchers with NGS or bioinformatics skills.
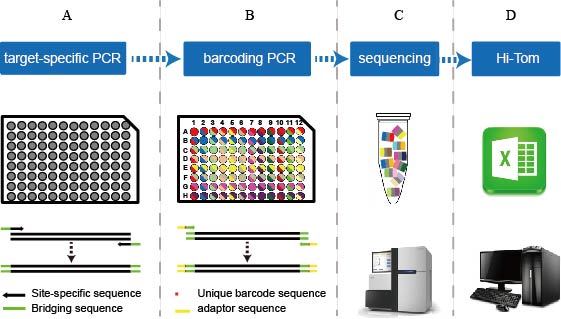
In this paper researchers developed Hi-TOM (available at http://www.hi-tom.net/hi-tom/), an online tool to track the mutations with precise percentage for multiple samples and multiple target sites. This paper also describes a next-generation sequencing (NGS) library construction strategy by fixing the bridge sequences and barcoding primers which are consistent with Hi-TOM software. Analysis of the samples from rice, hexaploid wheat and human cells reveals that the Hi-TOM tool has high reliability and sensitivity in tracking various mutations, especially complex chimeric mutations that frequently induced by genome editing. Hi-TOM does not require special design of barcode primers, cumbersome parameter configuration or additional data analysis. Thus, the streamlined NGS library construction and comprehensive result output make Hi-TOM particularly suitable for high-throughput identification of all types of mutations induced by CRISPR/Cas systems.
This study was supported by the National Key Research and Development Program of China (2017YFD0102002), the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, and the National Natural Science Foundation of China (31401363). More details are available on the link below: http://engine.scichina.com/doi/10.1007/s11427-018-9402-9
· Study Reveals How the Clock Component OsLUX Regulates Rice Heading
· Scientists Further Unravelled the Underlying Mechanism of Heading Date Control in Rice
· Improving the efficiency of prime editing in rice
· A New Infection Mechanism of Ustilaginoidea virens is Revealed
· Scientists Further Reveal the Infection Mechanism of Ustilaginoidea virens
CNRRI Today
Copyright © 2014- China National Rice Research Institute